
It codes information in genes.B M B 400, Part Three. DNA: resides in the nucleus. During protein synthesis, a ribosome moves along the mRNA template and, using aminoacyl-tRNAs, decodes the template nucleotide triplets to assemble a. With the element of each spanning two different sites on the ribosome The enzyme activity that catalyzes peptide bond formation has historically been referred to as peptidyl transferase and was widely assumed to be intrinsic to. Protein synthesis (Translation. Protein synthesis is the essential biological process, occurring inside the cell.
We will cover the material in that order, since that is the direction that information flows.A: Only twenty of the available amino acids are often present in proteins among. Recall the Central Dogma of molecular biology: DNA is transcribed into RNA, which is translated into protein. Overview of Part Three: The pathway of gene expression.
They are the end product of what's encoded in the genes and they perform all the functions in the cell. Protein: synthesized by ribosomes. Translation: ribosomes read off the mRNAs to make proteins. MRNAs are working copies of the gene. RNA: The mRNAs get transported out of the nucleus into the cytoplasm. Transcription: Inside the nucleus, the DNA genes get transcribed into RNA (messenger RNAs or mRNAs).
Degenerate because more than one codon codes for a given amino acid Non-overlapping because the 3 nucleotides that consist of one codon never serve as part of another codon There're no nucleotides in between Continuous because one codon follows right after another. Codons are continuous, non-overlapping and degenerate To do this: 3 nucleotide = 1 codon = 1 amino acid
mRNA composition and structure (RNA nucleotides, 5' cap, poly-A tail) Instead a protein called "release factor" comes along and terminates translation Unlike other codons, tRNA are not involved. Termination codon (UAG,UGA,UAA): signals the end of translation. Lies just downstream of the Shine Dalgarno sequence (Kozak sequence for eukaryotes)
The poly-A tail protects the 3' end of the mRNA from exonuclease degradation. This protects the 5' end from exonuclease degradation. The 5' cap is a modified nucleotide linked in a special way to the mRNA. It's the product of transcription and the template for translation.
Both tRNA (transfer RNA) and rRNA (ribosomal RNA) are products of transcription. tRNA, rRNA composition and structure (eg., RNA nucleotides) Stronger promoter, less DNA/histone methylation = more mRNA made More mRNA = more level of expression = more protein made Prokaryotic mRNAs don't have the 5' cap or polyA tail.
tRNA structure: clover leaf structure with anticodon at the tip, and the amino acid at the 3' tail. At the 3' end of the tRNA, the amino acid is attached to the 3'OH via an ester linkage. tRNA is made of nucleotides, many of which is modified for structural and functional reasons. RRNA makes up the ribosome, which is the enzyme responsible for translation. TRNA is responsible for bringing in the correct amino acid during translation.
Rho (ρ) dependent termination: a protein called the ρ factor travels along the synthesized RNA and bumps off the polymerase. Intrinsic termination: specific sequences called a termination site creates a stem-loop structure on the RNA that causes the RNA to slip off the template. Chain termination: there are 2 ways that transcription can terminate. RNA is made from the 5' to 3' direction.
Ribozymes = RNA enzymes = can have protein parts, but it's actually the RNA part that's doing the catalysis. Ribozymes, spliceosomes, small nuclear ribonucleoproteins (snRNPs), small nuclear RNAs (snRNAs) Alternate splicing = different ways you can do RNA splicing to make different end products RNA splicing: cut out introns, keep and stitch together the exons protect from degradation: 5' cap and 3' polyA added
Evolution: evolved from selfish/parasitic/mobile genetic elements Function: RNA splicing, alternate splicing, gene regulation Functional and evolutionary importance of introns snRNPs = RNA + protein = subunits that assemble into the spliceosomes and other RNA modification machinery Spliceosomes = machinery that does RNA splicing = composed of RNA and proteins = possibly a ribozyme
rRNA (ribosomal RNA): forms the ribosome. Link the correct amino acid to its corresponding mRNA codon through codon-anticodon interaction. tRNA (transfer RNA): contains the anticodon on the "tip" and the corresponding amino acid on the "tail".
Both subunits are needed for translation to occur and they come together in a hamburger fashion that sandwiches the mRNA and tRNAs in between. The small subunit is responsible for the recognizing mRNA and binds to the Shine-Dalgarno sequence on the mRNA (Kozak sequence for eukaryotes). The large subunit is responsible for the peptidyl transfer reaction. Ribosome has 2 subunits - the large and the small. Ribosome is the enzyme that catalyzes protein synthesis.
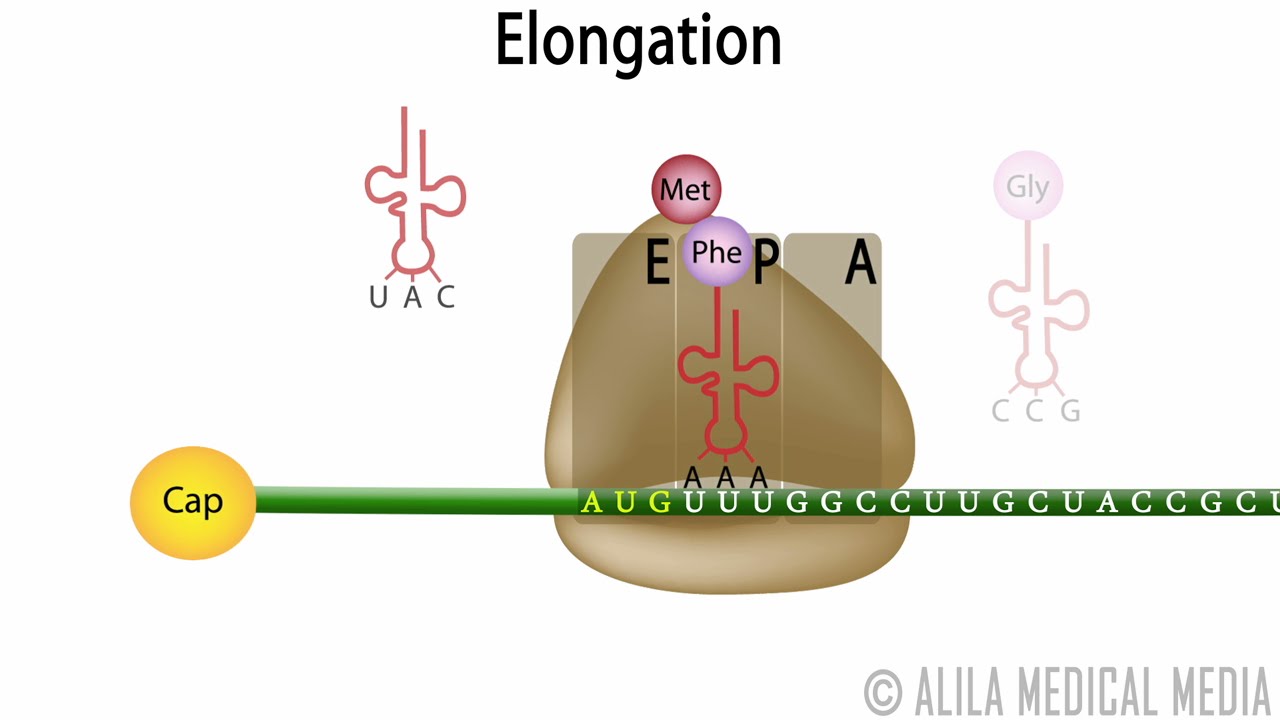
This includes mRNA, initiator tRNA (fmet), and the ribosome (initiation factors, and GTP aids in the formation of the initiation complex). The initiation complex is basically an assembly of everything needed to begin translation. Chain Initiation: To begin translation, you need to form the initiation complex.
Peptidyl transfer: attachment of the new amino acid to the existing chain in the P site. GTP and elongation factor required. Binding: new tRNA with its amino acid (tRNA+amino acid is called aminoacyl-tRNA) enters the A site. MRNA codons are read from the 5' to the 3' end. Chain Elongation: protein is made from the N terminus to the C terminus. The Shine-Dalgarno sequence is the "promoter" equivalent of translation for prokaryotes (Kozak sequence for eukaryotes).
The A site is now empty and ready for the binding of a new aminoacyl-tRNA to a new codon. The mRNA gets dragged along also - the codon that was in the A site is now in the P site after translocation. Translocation: the lone tRNA in the P site gets kicked off (E site), and the tRNA in the A site, along with the peptide chain attached to it, moves into the P site.
Peptide chain falls off, and then the whole translation complex falls apart. The peptide chain gets cleaved from the tRNA in the P site. Chain termination: When a stop codon is encountered, proteins called release factors, bound to GTP, come in and blocks the A site.


 0 kommentar(er)
0 kommentar(er)
